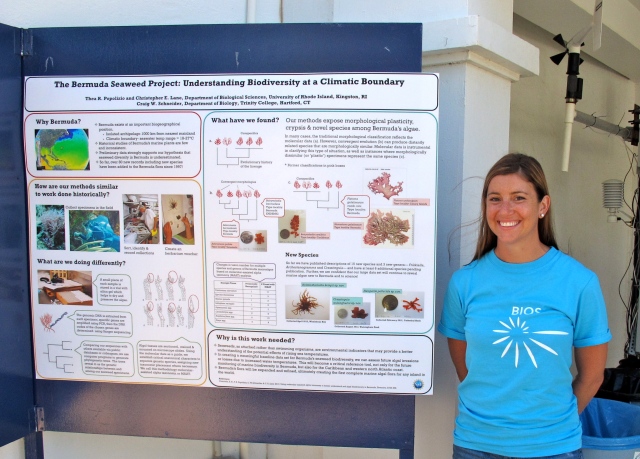
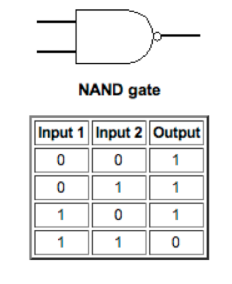
We all know that plants ‘eat’ through the process called photosynthesis, a chemical process where light energy and carbon dioxide are turned into carbohydrates, oxygen and water. In plants and algae, the process of photosynthesis occurs in the chloroplast, an organelle found within the individual cells. It has become widely accepted that the plant and algal chloroplast originated as a cyanobacteria that was engulfed by another single celled organism, resulting in a symbiotic relationship. After this primary ‘endosymbiotic event’ the original cyanobacteria (now chloroplast) transferred genes that are needed for its survival to the nucleus of the symbiont. The symbiont makes proteins needed to maintain the chloroplast, and in turns receives carbohydrates produced in the chloroplast for its own survival.

Illustration of the origin of the plant and algal chloroplast. Image from: http://biol10005.tumblr.com/post/32506576425/evolution-of-the-eukaryotic-cell
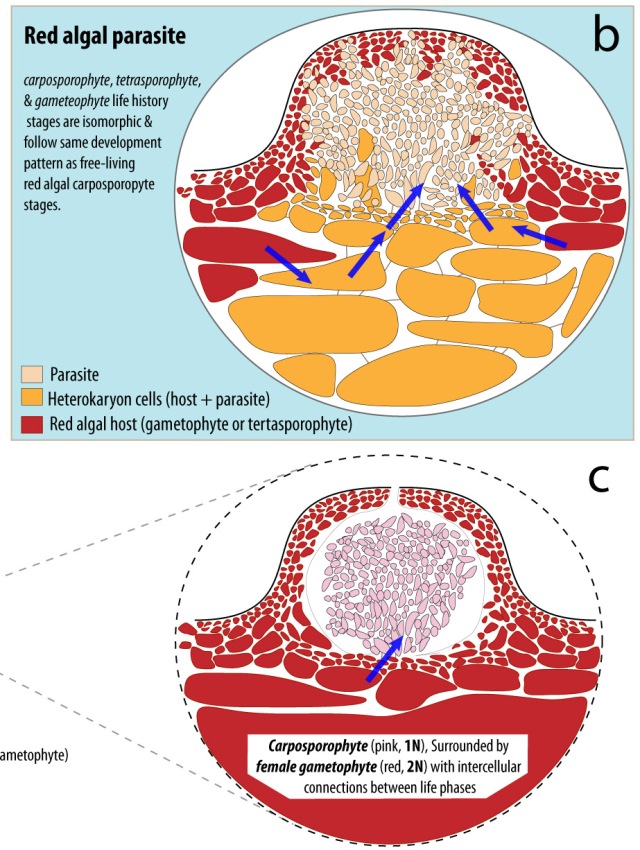
Plants and algae are not the only organisms that have found a way to photosynthesize. A few species of sacoglossan sea slugs manage to steal the chloroplasts from some of the species of algae that they eat and are able to maintain these ‘kleptoplasts’ for their own benefit. It has been known that these sea slugs are able to maintain the kleptoplasts for extended periods of time, enabling the slugs to survive during extended periods of starvation. However the mechanisms behind the longevity of the kleptoplasts remains elusive. Normal photosynthetic function requires the continual replacement of degraded proteins required for the chloroplast to photosynthesize. It has been hypothesized that the algal genes that manufacture these proteins have been transferred to the sea slugs, enabling the sea slugs to produce those proteins and maintain the kleptoplasts. However, the results from transcriptome and genome surveys conducted on a few of these sea slugs provided no evidence for the presence any genes or DNA of algal origin in the sea slug, adding further intrigue into the mechanism of kleptoplast retention (Wägele et al., 2011; Rumpho et al., 2011;Bhattacharya et al., 2013).
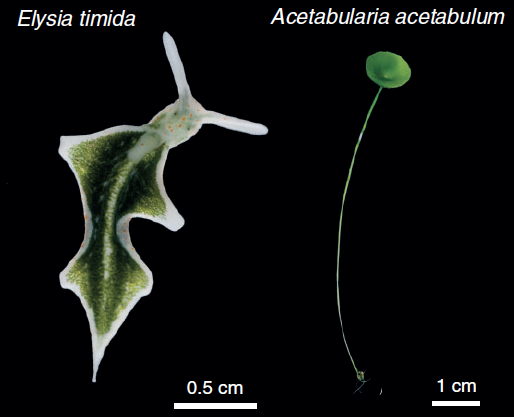
The sacoglossan sea slug, Elysia timida (left) and its primary food source and plastid contributor, Acetabularia acetabulum (right). Image from de Vries et al., 2014
Research by de Vries et al. (2014) examined chloroplast genome content from the algae Acetabularia acetabulum and Vaucheria litorea, the preferred food sources of the sea slugs, Elysia timida and Elysia chlorotica respectively. This study revealed that the chloroplasts encoded the genes tufA and ftsH, which are not present in the plastids of many land plant chloroplasts. Furthermore, after a month of starvation, transcripts of the plastid genes tufA, ftsH and psbA were all found in E. timida (de Vries et al., 2014) supporting the previous findings of tufA and ftsH transcripts from V. litorea plastids in E. chlorotica after two months of starvation. The presence of these genes are noteworthy since ftsH is essential for the maintenance of the photosystem II (a major part of the photosynthetic pathway), and tufA plays a role in enabling the protein from fstH to be made. It is also noteworthy that, while the sea slugs eat other algae including Bryopsis sp., the Bryopsis chloroplast is not retained by the sea slugs, and does not encode the ftsH gene.
This could mean that the presence of ftsH, tufA and other genes involved in maintaining the photosynthetic function are the key to long-term retention of algal chloroplasts in sea slugs. It is possible that the sea slugs are not actually providing any protein to the kleptoplast and are not actively maintaining the plastid. With this in mind it seems only logical that we conclude that these sacoglossan sea slugs have evolved into photosynthetic pirates, plundering algae for their plastids and digesting everything else in their wake.

Photosynthetic Sea Slug Pirate. Image from: http://javaape42.deviantart.com/art/Pirate-Slug-colored-380400956
References:
Bhattacharya D, Pelletreau KN, Price DC, Sarver KE, Rumpho ME. (2013) Genome analysis of Elysia chlorotica egg DNA provides no evidence for horizontal gene transfer into the germ line of this kleptoplastic mollusc. Mol Biol Evol 30:1843-1852.
De Vries J, Habicht J, Woehle C, Huang C, Christa G, Wägele H, Nickelsen J, Martin W Gould S. 2014. Is ftsH the key to plstid longevity in sacoglossan sea slugs? Genome Biol Evol 5:2540-2548
Rumpho ME, Pelletreau KN, Moustafa A, Bhattacharya D. 2011. The making of a photosynthetic animal. J Exp Biol 214:303-311.
Wägele H, Deusch O, Händeler K, Martin R, Schmitt V, Christa G, Pinzger B, Gould SB, Dagan T, Klussmann-Kolb, A, Martin W. 2011. Transcriptomic evidence that longevity of acquired plastids in the photosynthetic slugs Elysia timida and Plakobranchus ocellatus does not entail lateral transfer of algal nuclear genes. Mol Biol Evol 28:699–706.